Example: Bayesian quantification of parameter uncertainty:
Estimating the (Gaussian) posterior pdf of the coefficient parameter field in an elliptic PDE
In this example we tackle the problem of quantifying the uncertainty in the solution of an inverse problem governed by an elliptic PDE via the Bayesian inference framework. Hence, we state the inverse problem as a problem of statistical inference over the space of uncertain parameters, which are to be inferred from data and a physical model. The resulting solution to the statistical inverse problem is a posterior distribution that assigns to any candidate set of parameter fields our belief (expressed as a probability) that a member of this candidate set is the ``true’’ parameter field that gave rise to the observed data.
For simplicity, in what follows we give finite-dimensional expressions (i.e., after discretization of the parameter space) for the Bayesian formulation of the inverse problem.
Bayes’ Theorem:
The posterior probability distribution combines the prior pdf $\pi_{\text{prior}}(\m)$ over the parameter space, which encodes any knowledge or assumptions about the parameter space that we may wish to impose before the data are considered, with a likelihood pdf $\pi_{\text{like}}(\vec{d}{\text{obs}} \; | \; \m)$, which explicitly represents the probability that a given set of parameters $\m$ might give rise to the observed data $\vec{d}{\text{obs}} \in \mathbb{R}^m$, namely:
Note that infinite-dimensional analog of Bayes’ formula requires the use Radon-Nikodym derivatives instead of probability density functions.
Gaussian prior and noise:
The prior:
We consider a Gaussian prior with mean $\vec m_{\text prior}$ and covariance $\bf \Gamma_{\text{prior}}$. The covariance is given by the discretization of the inverse of differential operator $\mathcal{A}^{-2} = (-\gamma \Delta + \delta I)^{-2}$, where $\gamma$, $\delta > 0$ control the correlation length and the variance of the prior operator. This choice of prior ensures that it is a trace-class operator, guaranteeing bounded pointwise variance and a well-posed infinite-dimensional Bayesian inverse problem
The likelihood:
Here $\bf f$ is the parameter-to-observable map that takes a parameter vector $\m$ and maps it to the space observation vector $\data$.
The posterior:
The Gaussian approximation of the posterior: $\mathcal{N}(\vec{\map},\bf \Gamma_{\text{post}})$
The mean of this posterior distribution, $\vec{\map}$, is the parameter vector maximizing the posterior, and is known as the maximum a posteriori (MAP) point. It can be found by minimizing the negative log of the posterior, which amounts to solving a deterministic inverse problem) with appropriately weighted norms,
The posterior covariance matrix is then given by the inverse of the Hessian matrix of $\mathcal{J}$ at $\map$, namely
The generalized eigenvalue problem:
where $\matrix{\Lambda} = diag(\lambda_i) \in \mathbb{R}^{n\times n}$ contains the generalized eigenvalues and the columns of $\matrix V\in \mathbb R^{n\times n}$ the generalized eigenvectors such that $\matrix{V}^T \prcov^{-1} \matrix{V} = \matrix{I}$.
Randomized eigensolvers to construct the approximate spectral decomposition:
When the generalized eigenvalues ${\lambda_i}$ decay rapidly, we can extract a low-rank approximation of $\Hmisfit$ by retaining only the $r$ largest eigenvalues and corresponding eigenvectors,
Here, $\matrix{V}_r \in \mathbb{R}^{n\times r}$ contains only the $r$ generalized eigenvectors of $\Hmisfit$ that correspond to the $r$ largest eigenvalues, which are assembled into the diagonal matrix $\matrix{\Lambda}_r = \diag (\lambda_i) \in \mathbb{R}^{r \times r}$.
The approximate posterior covariance:
Using the Sherman–Morrison–Woodbury formula, we write
where $\matrix{D}_r :=\diag(\lambda_i/(\lambda_i+1)) \in \mathbb{R}^{r\times r}$. The last term in this expression captures the error due to truncation in terms of the discarded eigenvalues; this provides a criterion for truncating the spectrum, namely that $r$ is chosen such that $\lambda_r$ is small relative to 1.
Therefore we can approximate the posterior covariance as
Drawing samples from a Gaussian distribution with covariance $\H^{-1}$
Let $\bf x$ be a sample for the prior distribution, i.e. $\bf x \sim \mathcal{N}({\bf 0}, \prcov)$, then, using the low rank approximation of the posterior covariance, we compute a sample ${\bf v} \sim \mathcal{N}({\bf 0}, \H^{-1})$ as
This tutorial shows:
- description of the inverse problem (the forward problem, the prior, and the misfit functional)
- convergence of the inexact Newton-CG algorithm
- low-rank-based approximation of the posterior covariance (built on a low-rank approximation of the Hessian of the data misfit)
- how to construct the low-rank approximation of the Hessian of the data misfit
- how to apply the inverse and square-root inverse Hessian to a vector efficiently
- samples from the Gaussian approximation of the posterior
Goals:
By the end of this notebook, you should be able to:
- understand the Bayesian inverse framework
- visualise and understand the results
- modify the problem and code
Mathematical tools used:
- Finite element method
- Derivation of gradiant and Hessian via the adjoint method
- inexact Newton-CG
- Armijo line search
- Bayes’ formula
- randomized eigensolvers
List of software used:
- FEniCS, a parallel finite element element library for the discretization of partial differential equations
- PETSc, for scalable and efficient linear algebra operations and solvers
- Matplotlib, A great python package that I used for plotting many of the results
- Numpy, A python package for linear algebra. While extensive, this is mostly used to compute means and sums in this notebook.
1. Load modules
from __future__ import absolute_import, division, print_function
import dolfin as dl
import math
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import sys
sys.path.append( "../hippylib" )
from hippylib import *
sys.path.append( "../hippylib/tutorial" )
import nb
import logging
logging.getLogger('FFC').setLevel(logging.WARNING)
logging.getLogger('UFL').setLevel(logging.WARNING)
dl.set_log_active(False)
np.random.seed(seed=1)
2. Generate the true parameter
This function generates a random field with a prescribed anysotropic covariance function.
def true_model(prior):
noise = dl.Vector()
prior.init_vector(noise,"noise")
noise_size = noise.array().shape[0]
noise.set_local( np.random.randn( noise_size ) )
mtrue = dl.Vector()
prior.init_vector(mtrue, 0)
prior.sample(noise,mtrue)
return mtrue
3. Set up the mesh and finite element spaces
We compute a two dimensional mesh of a unit square with nx by ny elements. We define a P2 finite element space for the state and adjoint variable and P1 for the parameter.
ndim = 2
nx = 64
ny = 64
mesh = dl.UnitSquareMesh(nx, ny)
Vh2 = dl.FunctionSpace(mesh, 'Lagrange', 2)
Vh1 = dl.FunctionSpace(mesh, 'Lagrange', 1)
Vh = [Vh2, Vh1, Vh2]
print("Number of dofs: STATE={0}, PARAMETER={1}, ADJOINT={2}".format(
Vh[STATE].dim(), Vh[PARAMETER].dim(), Vh[ADJOINT].dim()) )
Number of dofs: STATE=16641, PARAMETER=4225, ADJOINT=16641
4. Set up the forward problem
To set up the forward problem we use the PDEVariationalProblem class, which requires the following inputs
- the finite element spaces for the state, parameter, and adjoint variables
Vh - the pde in weak form
pde_varf - the boundary conditions
bcfor the forward problem andbc0for the adjoint and incremental problems.
The PDEVariationalProblem class offer the following functionality:
- solving the forward/adjoint and incremental problems
- evaluate first and second partial derivative of the forward problem with respect to the state, parameter, and adojnt variables.
def u_boundary(x, on_boundary):
return on_boundary and ( x[1] < dl.DOLFIN_EPS or x[1] > 1.0 - dl.DOLFIN_EPS)
u_bdr = dl.Expression("x[1]", degree=1)
u_bdr0 = dl.Constant(0.0)
bc = dl.DirichletBC(Vh[STATE], u_bdr, u_boundary)
bc0 = dl.DirichletBC(Vh[STATE], u_bdr0, u_boundary)
f = dl.Constant(0.0)
def pde_varf(u,m,p):
return dl.exp(m)*dl.inner(dl.nabla_grad(u), dl.nabla_grad(p))*dl.dx - f*p*dl.dx
pde = PDEVariationalProblem(Vh, pde_varf, bc, bc0, is_fwd_linear=True)
4. Set up the prior
To obtain the synthetic true paramter $m_{\rm true}$ we generate a realization from the prior distribution. Here we assume a Gaussian prior with zero average and covariance matrix $\mathcal{C} = \mathcal{A}^{-2}$, where $\mathcal{A}$ is a differential operator of the form
Here $\Theta$ is an s.p.d. anisotropic tensor of the form
gamma = .1
delta = .5
anis_diff = dl.Expression(code_AnisTensor2D, degree=1)
anis_diff.theta0 = 2.
anis_diff.theta1 = .5
anis_diff.alpha = math.pi/4
prior = BiLaplacianPrior(Vh[PARAMETER], gamma, delta, anis_diff )
mtrue = true_model(prior)
print("Prior regularization: (delta_x - gamma*Laplacian)^order: delta={0}, gamma={1}, order={2}".format(
delta, gamma,2) )
objs = [dl.Function(Vh[PARAMETER],mtrue), dl.Function(Vh[PARAMETER],prior.mean)]
mytitles = ["True Parameter", "Prior mean"]
nb.multi1_plot(objs, mytitles)
plt.show()
model = Model(pde,prior, misfit)
Prior regularization: (delta_x - gamma*Laplacian)^order: delta=0.5, gamma=0.1, order=2
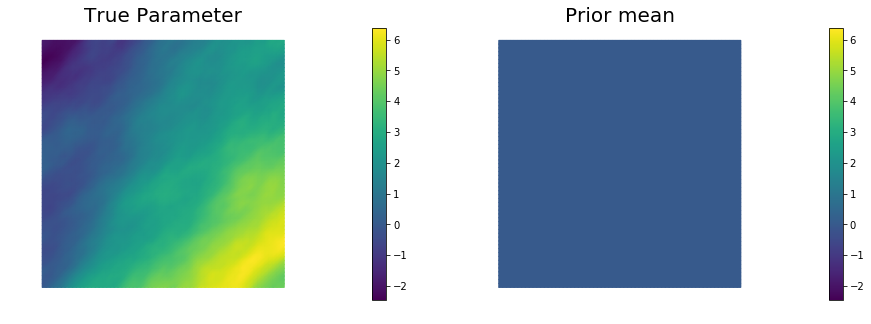
5. Set up the misfit functional and generate synthetic observations
To setup the observation operator, we generate ntargets random locations where to evaluate the value of the state.
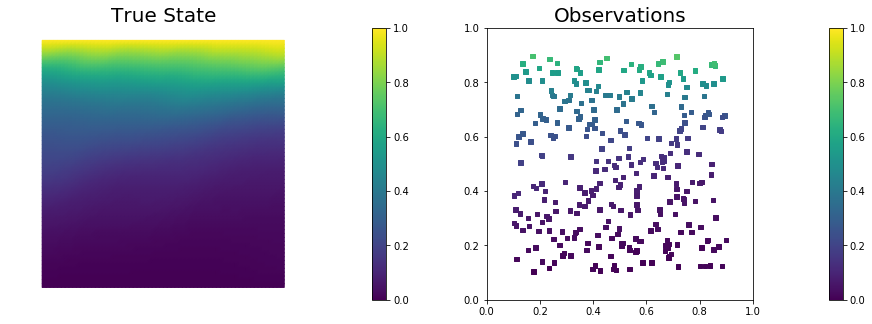
To generate the synthetic observation, we first solve the forward problem using the true parameter $m_{\rm true}$. Synthetic observations are obtained by perturbing the state variable at the observation points with a random Gaussian noise. rel_noise is the signal to noise ratio.
ntargets = 300
rel_noise = 0.005
targets = np.random.uniform(0.1,0.9, [ntargets, ndim] )
print("Number of observation points: {0}".format(ntargets))
misfit = PointwiseStateObservation(Vh[STATE], targets)
utrue = pde.generate_state()
x = [utrue, mtrue, None]
pde.solveFwd(x[STATE], x, 1e-9)
misfit.B.mult(x[STATE], misfit.d)
MAX = misfit.d.norm("linf")
noise_std_dev = rel_noise * MAX
randn_perturb(misfit.d, noise_std_dev)
misfit.noise_variance = noise_std_dev*noise_std_dev
vmax = max( utrue.max(), misfit.d.max() )
vmin = min( utrue.min(), misfit.d.min() )
plt.figure(figsize=(15,5))
nb.plot(dl.Function(Vh[STATE], utrue), mytitle="True State", subplot_loc=121, vmin=vmin, vmax=vmax)
nb.plot_pts(targets, misfit.d, mytitle="Observations", subplot_loc=122, vmin=vmin, vmax=vmax)
plt.show()
Number of observation points: 300
6. Set up the model and test gradient and Hessian
The model is defined by three component:
- the
PDEVariationalProblempdewhich provides methods for the solution of the forward problem, adjoint problem, and incremental forward and adjoint problems. - the
Priorpriorwhich provides methods to apply the regularization (precision) operator to a vector or to apply the prior covariance operator (i.e. to solve linear system with the regularization operator) - the
Misfitmisfitwhich provides methods to compute the cost functional and its partial derivatives with respect to the state and parameter variables.
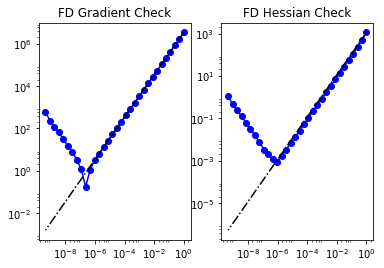
To test gradient and the Hessian of the model we use forward finite differences.
model = Model(pde, prior, misfit)
m0 = dl.interpolate(dl.Expression("sin(x[0])", degree=5), Vh[PARAMETER])
modelVerify(model, m0.vector(), 1e-12)
(yy, H xx) - (xx, H yy) = 3.67659462287e-14
7. Compute the MAP point
We used the globalized Newtown-CG method to compute the MAP point.
m0 = prior.mean.copy()
solver = ReducedSpaceNewtonCG(model)
solver.parameters["rel_tolerance"] = 1e-9
solver.parameters["abs_tolerance"] = 1e-12
solver.parameters["max_iter"] = 25
solver.parameters["inner_rel_tolerance"] = 1e-15
solver.parameters["c_armijo"] = 1e-4
solver.parameters["GN_iter"] = 5
x = solver.solve(m0)
if solver.converged:
print("\nConverged in ", solver.it, " iterations.")
else:
print("\nNot Converged")
print("Termination reason: ", solver.termination_reasons[solver.reason])
print("Final gradient norm: ", solver.final_grad_norm)
print("Final cost: ", solver.final_cost)
plt.figure(figsize=(15,5))
nb.plot(dl.Function(Vh[STATE], x[STATE]), subplot_loc=121,mytitle="State")
nb.plot(dl.Function(Vh[PARAMETER], x[PARAMETER]), subplot_loc=122,mytitle="Parameter")
plt.show()
It cg_it cost misfit reg (g,da) ||g||L2 alpha tolcg
1 1 2.413091e+04 2.413027e+04 6.399116e-01 -1.939374e+06 2.315299e+06 1.000000e+00 5.000000e-01
2 1 5.042246e+03 5.041320e+03 9.257374e-01 -3.866980e+04 2.942004e+05 1.000000e+00 3.564660e-01
3 4 1.127941e+03 1.126491e+03 1.450689e+00 -8.429483e+03 6.234143e+04 1.000000e+00 1.640910e-01
4 1 8.391944e+02 8.377019e+02 1.492567e+00 -5.800344e+02 3.673645e+04 1.000000e+00 1.259636e-01
5 17 7.518823e+02 7.431716e+02 8.710769e+00 -6.167742e+04 1.840903e+04 3.906250e-03 8.916859e-02
6 3 5.053155e+02 4.964618e+02 8.853717e+00 -4.657349e+02 2.426426e+04 1.000000e+00 1.023717e-01
7 10 2.136538e+02 2.023889e+02 1.126493e+01 -6.176476e+02 1.133936e+04 1.000000e+00 6.998272e-02
8 2 2.022320e+02 1.909626e+02 1.126942e+01 -2.272049e+01 7.516712e+03 1.000000e+00 5.697843e-02
9 78 1.856770e+02 1.476523e+02 3.802472e+01 -1.567853e+02 3.162018e+03 5.000000e-01 3.695547e-02
10 28 1.687066e+02 1.400432e+02 2.866333e+01 -6.004973e+02 2.551508e+03 6.250000e-02 3.319671e-02
11 4 1.610230e+02 1.323686e+02 2.865438e+01 -1.518996e+01 4.053085e+03 1.000000e+00 4.183978e-02
12 58 1.502097e+02 1.255140e+02 2.469572e+01 -5.807913e+01 1.414695e+03 2.500000e-01 2.471883e-02
13 8 1.457503e+02 1.207485e+02 2.500179e+01 -9.204138e+00 1.960018e+03 1.000000e+00 2.909555e-02
14 45 1.404043e+02 1.136082e+02 2.679611e+01 -1.198721e+01 8.921054e+02 1.000000e+00 1.962929e-02
15 6 1.400080e+02 1.132007e+02 2.680729e+01 -7.965490e-01 8.152519e+02 1.000000e+00 1.876473e-02
16 70 1.399017e+02 1.132745e+02 2.662723e+01 -2.117833e-01 1.251631e+02 1.000000e+00 7.352495e-03
17 32 1.399015e+02 1.132692e+02 2.663233e+01 -3.238605e-04 9.485365e+00 1.000000e+00 2.024061e-03
18 91 1.399015e+02 1.132692e+02 2.663239e+01 -6.580644e-06 6.295916e-01 1.000000e+00 5.214659e-04
Converged in 18 iterations.
Termination reason: Norm of the gradient less than tolerance
Final gradient norm: 0.000647377811664
Final cost: 139.901546319
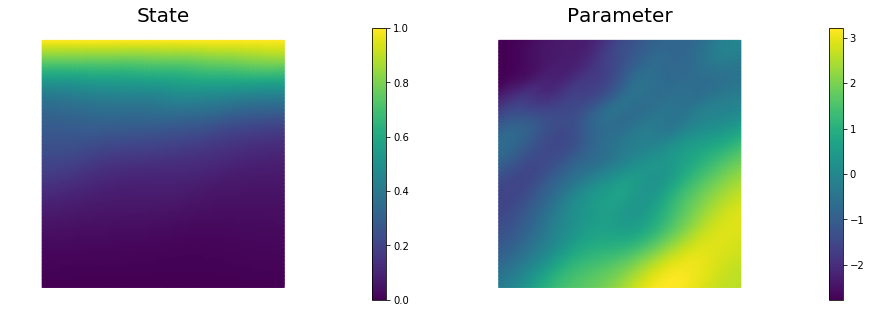
8. Compute the low rank Gaussian approximation of the posterior
We used the double pass algorithm to compute a low-rank decomposition of the Hessian Misfit. In particular, we solve
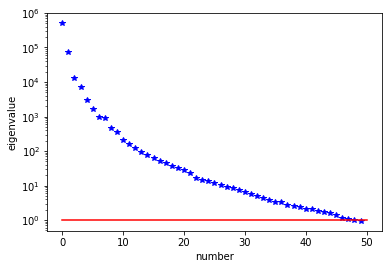
The Figure shows the largest k generalized eigenvectors of the Hessian misfit. The effective rank of the Hessian misfit is the number of eigenvalues above the red line ($y=1$). The effective rank is independent of the mesh size.
model.setPointForHessianEvaluations(x)
Hmisfit = ReducedHessian(model, solver.parameters["inner_rel_tolerance"], gauss_newton_approx=False, misfit_only=True)
k = 50
p = 20
print("Single/Double Pass Algorithm. Requested eigenvectors: {0}; Oversampling {1}.".format(k,p))
Omega = np.random.randn(x[PARAMETER].array().shape[0], k+p)
lmbda, V = doublePassG(Hmisfit, prior.R, prior.Rsolver, Omega, k)
posterior = GaussianLRPosterior(prior, lmbda, V)
posterior.mean = x[PARAMETER]
plt.plot(range(0,k), lmbda, 'b*', range(0,k+1), np.ones(k+1), '-r')
plt.yscale('log')
plt.xlabel('number')
plt.ylabel('eigenvalue')
nb.plot_eigenvectors(Vh[PARAMETER], V, mytitle="Eigenvector", which=[0,1,2,5,10,15])
Single/Double Pass Algorithm. Requested eigenvectors: 50; Oversampling 20.
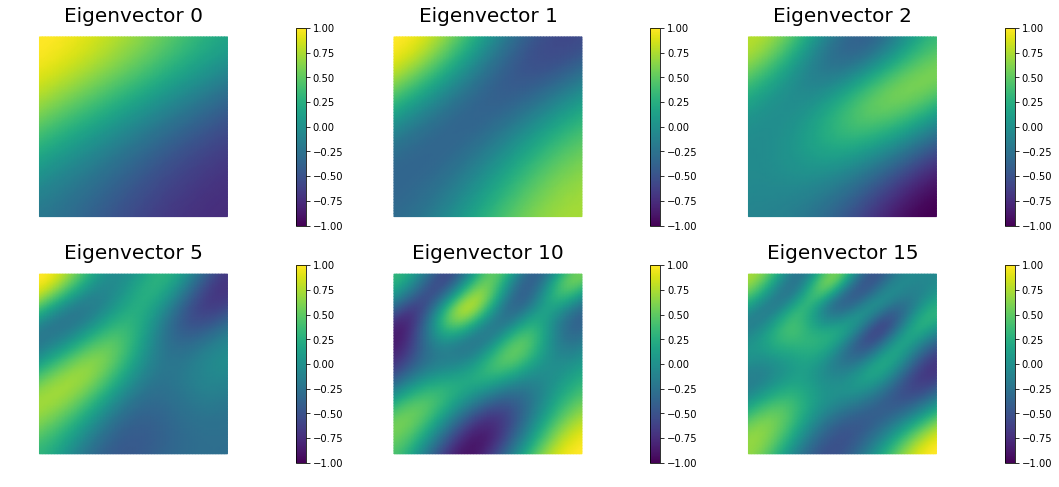
9. Prior and posterior pointwise variance fields
compute_trace = True
if compute_trace:
post_tr, prior_tr, corr_tr = posterior.trace(method="Estimator", tol=5e-2, min_iter=20, max_iter=2000)
print("Posterior trace {0:5e}; Prior trace {1:5e}; Correction trace {2:5e}".format(post_tr, prior_tr, corr_tr))
post_pw_variance, pr_pw_variance, corr_pw_variance = posterior.pointwise_variance("Exact")
objs = [dl.Function(Vh[PARAMETER], pr_pw_variance),
dl.Function(Vh[PARAMETER], post_pw_variance)]
mytitles = ["Prior variance", "Posterior variance"]
nb.multi1_plot(objs, mytitles, logscale=True)
plt.show()
Posterior trace 4.331423e+00; Prior trace 5.794617e+00; Correction trace 1.463193e+00
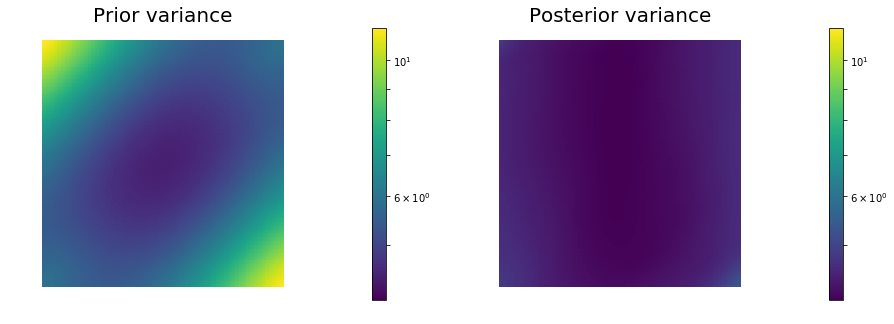
10. Generate samples from Prior and Posterior
nsamples = 5
noise = dl.Vector()
posterior.init_vector(noise,"noise")
noise_size = noise.array().shape[0]
s_prior = dl.Function(Vh[PARAMETER], name="sample_prior")
s_post = dl.Function(Vh[PARAMETER], name="sample_post")
pr_max = 2.5*math.sqrt( pr_pw_variance.max() ) + prior.mean.max()
pr_min = -2.5*math.sqrt( pr_pw_variance.max() ) + prior.mean.min()
ps_max = 2.5*math.sqrt( post_pw_variance.max() ) + posterior.mean.max()
ps_min = -2.5*math.sqrt( post_pw_variance.max() ) + posterior.mean.min()
for i in range(nsamples):
noise.set_local( np.random.randn( noise_size ) )
posterior.sample(noise, s_prior.vector(), s_post.vector())
plt.figure(figsize=(15,5))
nb.plot(s_prior, subplot_loc=121, mytitle="Prior sample", vmin=pr_min, vmax=pr_max)
nb.plot(s_post, subplot_loc=122, mytitle="Posterior sample", vmin=ps_min, vmax=ps_max)
plt.show()
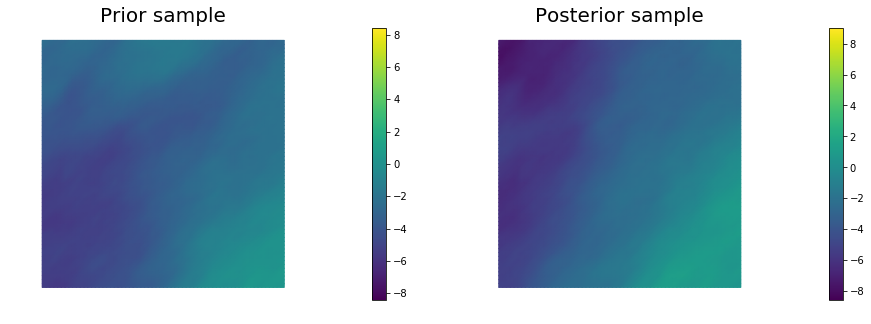
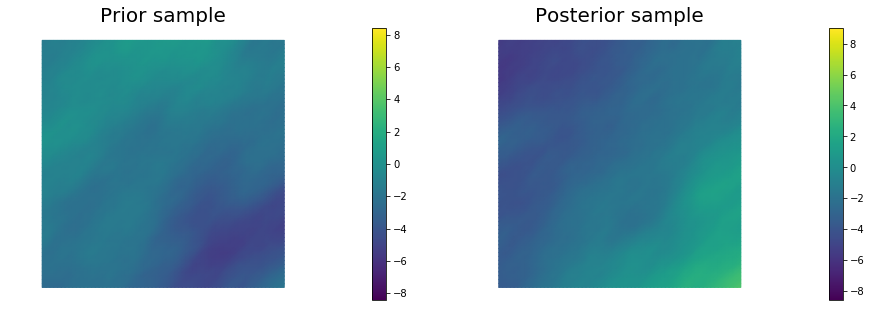
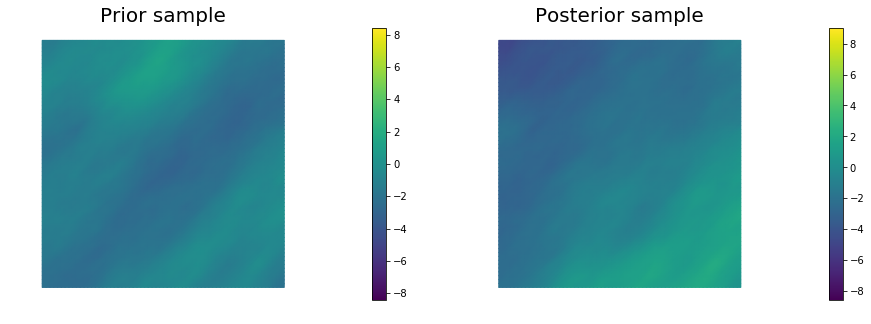
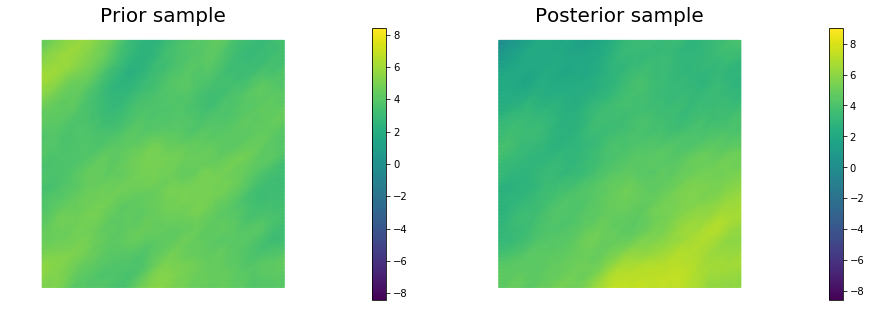
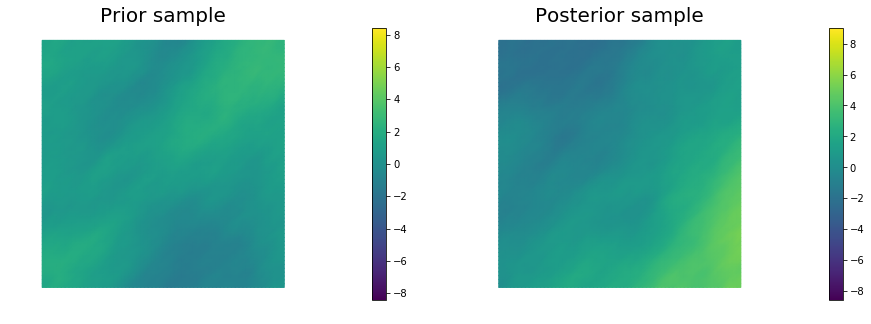
Copyright (c) 2016-2017, The University of Texas at Austin & University of California, Merced. All Rights reserved. See file COPYRIGHT for details.
This file is part of the hIPPYlib library. For more information and source code availability see https://hippylib.github.io.
hIPPYlib is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License (as published by the Free Software Foundation) version 2.0 dated June 1991.