An illustrative example
1D Inverse Heat Equation
An illustrative example is the inversion for the initial condition for a one-dimensional heat equation.
Specifically, we consider a rod of lenght $L$, and we are interested in reconstructing the initial temperature profile $m$ given some noisy measurements $d$ of the temperature profile at a later time $T$.
Forward problem
Given
- the initial temperature profile $u(x,0) = m(x)$,
- the termal diffusivity $k$,
- a prescribed temperature $u(0,t) = u(L,t) = 0$ at the extremities of the rod;
solve the heat equation
and observe the temperature at the final time $T$:
Analytical solution to the forward problem.
Verify that if
then
is the unique solution to the heat equation.
Inverse problem
Given the forward model $\mathcal{F}$ and a noisy measurament $d$ of the temperature profile at time $T$, find the initial temperature profile $m$ such that
Ill-posedness of the inverse problem
Consider a perturbation
where $\varepsilon > 0$ and $n = 1, 2, 3, \ldots$.
Then, by linearity of the forward model $\mathcal{F}$, the corresponding perturbation $\delta d(x) = \mathcal{F}(m + \delta m) - \mathcal{F}(m)$ is
which converges to zero as $n \rightarrow +\infty$.
Hence the ratio between $\delta m$ and $\delta d$ can become arbitrary large, which shows that the stability requirement for well-posedness can not be satisfied.
Discretization
To discretize the problem, we use finite differences in space and Implicit Euler in time.
Semidiscretization in space
We divide the $[0, L]$ interval in $n_x$ subintervals of the same lenght $h = \frac{L}{n_x}$, and we denote with $u_j(t) := u( jh, t)$ the value of the temperature at point $x_j = jh$ and time $t$.
We then use a centered finite difference approximation of the second derivative in space and write
with the boundary condition $u_0(t) = u_{n_x}(t) = 0$.
By letting
be the vector collecting the values of the temperature $u$ at the points $x_j = j\,h$, we then write the system of ordinary differential equations (ODEs):
where $K \in \mathbb{R}^{(n_x-1) \times (n_x-1)}$ is the tridiagonal matrix given by
Time discretization
We subdivide the time interval $(0, T]$ in $n_t$ time step of size $\Delta t = \frac{T}{n_t}$. By letting $\mathbf{u}^{(i)} = \mathbf{u}(i\,\Delta t)$ denote the discretized temperature profile at time $t_i = i\,\Delta t$, the Implicit Euler scheme reads
After simple algebraic manipulations and exploiting the initial condition $u(x,0) = m(x)$, we then obtain
or equivalently
In the code below, the function assembleMatrix generates the finite difference matrix $\left( I + \Delta t\, K \right)$ and the function solveFwd evaluates the forward model
import numpy as np
import scipy.sparse as sp
import scipy.sparse.linalg as la
import matplotlib.pyplot as plt
%matplotlib inline
def plot(f, style, **kwargs):
x = np.linspace(0., L, nx+1)
f_plot = np.zeros_like(x)
f_plot[1:-1] = f
plt.plot(x,f_plot, style, **kwargs)
def assembleMatrix(n):
diagonals = np.zeros((3, n)) # 3 diagonals
diagonals[0,:] = -1.0/h**2
diagonals[1,:] = 2.0/h**2
diagonals[2,:] = -1.0/h**2
K = k*sp.spdiags(diagonals, [-1,0,1], n,n)
M = sp.spdiags(np.ones(n), 0, n,n)
return M + dt*K
def solveFwd(m):
A = assembleMatrix(m.shape[0])
u_old = m.copy()
for i in np.arange(nt):
u = la.spsolve(A, u_old)
u_old[:] = u
return u
A naive solution to the inverse problem
If $\mathcal{F}$ is invertible a naive solution to the inverse problem $\mathcal{F} m = d$ is simply to set
The function naiveSolveInv computes the solution of the discretized inverse problem $\mathbf{m} = F^{-1} \mathbf{d}$ as
The code below shows that:
- for a very coarse mesh (
nx = 20) and no measurement noise (noise_std_dev = 0.0) the naive solution is quite good - for a finer mesh (
nx = 100) and/or even small measurement noise (noise_std_dev = 1e-4) the naive solution is garbage
def naiveSolveInv(d):
A = assembleMatrix(d.shape[0])
p_i = d.copy()
for i in np.arange(nt):
p = A*p_i
p_i[:] = p
return p
T = 1.0
L = 1.0
k = 0.005
nx = 20
nt = 100
noise_std_dev = 1e-4
h = L/float(nx)
dt = T/float(nt)
x = np.linspace(0.+h, L-h, nx-1) #place nx-1 equispace point in the interior of [0,L] interval
#m_true = np.power(.5,-36)*np.power(x,20)*np.power(1. - x, 16) #smooth true initial condition
m_true = 0.5 - np.abs(x-0.5) #initial condition with a corner
u_true = solveFwd(m_true)
d = u_true + noise_std_dev*np.random.randn(u_true.shape[0])
m = naiveSolveInv(d)
plot(m_true, "-r", label = 'm_true')
plot(m, "-b", label = 'm')
plt.legend()
plt.show()
plot(u_true, "-b", label = 'u(T)')
plot(d, "og", label = 'd')
plt.legend()
plt.show()
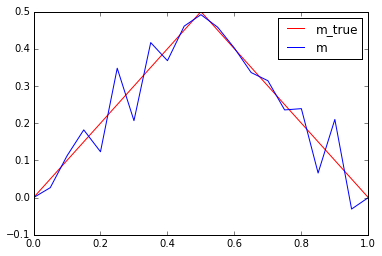
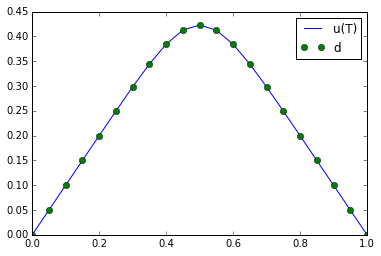
Why does the naive solution fail?
Let $v_n = \sqrt{\frac{2}{L}} \sin\left( n \, \frac{\pi}{L} x \right)$ with $n=1,2,3, \ldots$, then we have that
Note 1:
- Large eigenvalues $\lambda_n$ corresponds to smooth eigenfunctions $v_n$;
- Small eigenvalues $\lambda_n$ corresponds to oscillatory eigenfuctions $v_n$.
The figure below shows that the eigenvalues $\lambda_n$ decays extremely fast, that is the matrix $F$, discretization of the forward model $\mathcal{F}$, is extremely ill conditioned.
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
T = 1.0
L = 1.0
k = 0.005
i = np.arange(1,50)
lambdas = np.exp(-k*T*np.power(np.pi/L*i,2))
plt.semilogy(i, lambdas, 'ob')
plt.xlabel('i')
plt.ylabel('lambda_i')
plt.show()
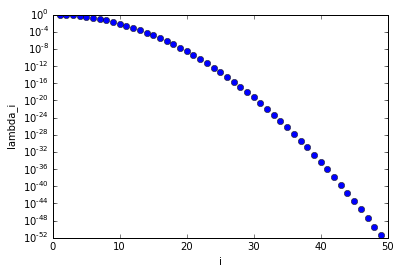
Note 2: The functions $v_n$, $n=1,2,3, \ldots$,form an orthonormal basis of $L^2([0,1])$.
That is, every function $f \in L^2([0,1])$ can be written as
Consider now the noisy problem
where
- $d$ is the data (noisy measurements)
- $\eta$ is the noise: $\eta(x) = \sum_{n=1}^\infty \eta_n v_n(x)$
- $m_{\rm true}$ is the true value of the parameter that generated the data
- $\mathcal{F}$ is the forward heat equation
Then, the naive solution to the inverse problem $\mathcal{F}m = d$ is
If the coefficients $\eta_n = \int_0^1 \eta(x) \, v_n(x) \, dx$ do not decay sufficiently fast with respect to the eigenvalues $\lambda_n$, then the naive solution is unstable.
This implies that oscillatory components can not reliably be reconstructed from noisy data since they correspond to small eigenvalues.
Regularization by filtering
Remedy is to dampen the terms corresponding to small eigenvalues and write
where
- $\delta_n = \int_0^1 d(x) \, v_n(x) \, dx$ denotes the coefficients of the data $d$ in the basis ;
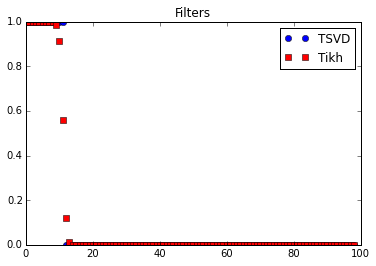
- $\omega( \lambda_n^2)$ is a filter function that allows to drop/stabilize the terms corresponding to small $\lambda_n$.
Truncated Singular Value Decomposition:
where $\alpha$ is a regularization parameter.
Then, we have
where $N$ is largest index such that $\lambda_n^2 \geq \alpha$ (assuming that $\lambda_n$ are sorted in a decreasing order).
Tikhonov filter:
where $\alpha$ is a regularization parameter.
NOTE: $\omega_\alpha( \lambda^2 )$ is close to $1$ when $\lambda \gg \alpha$, close to 0 when $\lambda \ll \alpha$).
Then we have
def computeEigendecomposition(n):
## Compute F as a dense matrix
F = np.zeros((n,n))
m_i = np.zeros(n)
for i in np.arange(n):
m_i[i] = 1.0
F[:,i] = solveFwd(m_i)
m_i[i] = 0.0
## solve the eigenvalue problem
lmbda, U = np.linalg.eigh(F)
## sort eigenpairs in decreasing order
lmbda[:] = lmbda[::-1]
lmbda[lmbda < 0.] = 0.0
U[:] = U[:,::-1]
return lmbda, U
## Setup the problem
T = 1.0
L = 1.0
k = 0.005
nx = 100
nt = 100
noise_std_dev = 0.0#1e-3
h = L/float(nx)
dt = T/float(nt)
## Compute the data d by solving the forward model
x = np.linspace(0.+h, L-h, nx-1)
#m_true = np.power(.5,-36)*np.power(x,20)*np.power(1. - x, 16)
m_true = 0.5 - np.abs(x-0.5)
u_true = solveFwd(m_true)
d = u_true + noise_std_dev*np.random.randn(u_true.shape[0])
## Compute eigenvector and eigenvalues of the discretized forward operator
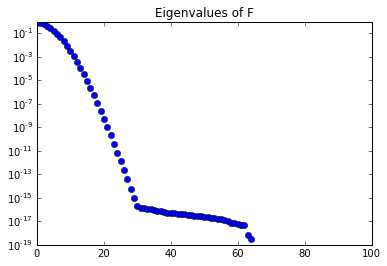
lmbda, U = computeEigendecomposition(nx-1)
plt.semilogy(lmbda, 'ob')
plt.title("Eigenvalues of F")
plt.show()
# Show filters
alpha = 1.e-6
plt.plot(lmbda*lmbda >= alpha, 'ob', label="TSVD")
plt.plot(lmbda*lmbda/(lmbda*lmbda + alpha), 'sr', label="Tikh")
plt.legend()
plt.title("Filters")
plt.show()
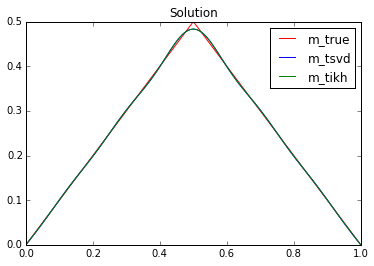
# Compute TSVD and Tikh sol
lmbda_inv = np.zeros_like(lmbda)
lmbda_inv[lmbda >= np.sqrt(alpha)] = lmbda[lmbda >= np.sqrt(alpha)]**(-1)
m_tsvd = np.dot(U, lmbda_inv*np.dot(U.T, d))
den = lmbda*lmbda+alpha
m_tikh = np.dot(U, (lmbda/den)*np.dot(U.T, d))
plot(m_true, "-r", label = 'm_true')
plot(m_tsvd, "-b", label = 'm_tsvd')
plot(m_tikh, "-g", label = 'm_tikh')
plt.title("Solution")
plt.legend()
plt.show()
Copyright © 2017, The University of Texas at Austin & University of California, Merced. All Rights reserved. See file COPYRIGHT for details.
This file is part of the hIPPYlib library. For more information and source code availability see https://hippylib.github.io.
hIPPYlib is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License (as published by the Free Software Foundation) version 2.0 dated June 1991.